BD2K: Big Data to Knowledge
The overarching aim of our program is to make Big Data seem smaller to every individual researcher.
We will accomplish this by attacking the problem on several fronts:
- look for redundancies and overlap that will allow us to decrease the size of Big Data.
- organize big data by high quality, standard ontologies linked through a common upper level ontology that will allow us more structured ways to slice and interrogate the Big Data.
- create new algorithms and models for handling Big Data across data types and data sources.
Together these will serve as an Electron Microscope for Big Data allowing researchers to focus on fine grain details from within indexed Big Data.
We bring together biomedical informatics, medicine, ontology, genetics, computer science, biology, biochemistry, biostatistics, epidemiology and Industrial Engineering experts to collaborate on Big Data Science Research. This collaborative environment is what is needed to advance the science. We propose to deliver an essential piece of the Big Data puzzle and work collaboratively with the other cooperative agreement awardees toward a principal set of approaches to handling, organizing and analyzing complex Big Data.
NY Genome Medicine Center
New York State has recently funded a collaboration between the University at Buffalo and the NY Genome Center to sequence a large number of individual’s genomes and to perform clinical genomic research. Biomedical Informatics is a central part of this project as expressed in a press release by UB President Satish K. Tripathi:
If our Big Data Center grant is funded it will synergize with and benefit from the work to be performed and the sequences and clinical data acquired by the NY Genome Medicine Center.
The National Center for Ontological Research (NCOR) was established in Buffalo in 2005 with the goal of advancing the quality of ontological research and development and of establishing tools and measures for ontology evaluation and quality assurance. NCOR draws on the expertise of ontologists associated with the University at Buffalo and of their collaborators in scientific, commercial and government institutions throughout the world.
NCOR serves as a vehicle to coordinate, enhance, publicize, and seek funding for ontological research activities. It provides coordination, infrastructure, and independent review to organizations employing ontologies in fields such as defense and intelligence, management, healthcare and biomedical sciences.
It provides researchers working in ontology-related areas with specialized support in seeking external funding and in assembling collaborative, interdisciplinary teams both nationally and internationally. It provides consultant services for ontology projects especially in the defense and security fields and in health care and biomedical informatics. NCOR also engages in training and outreach endeavors that are designed to broaden the range of institutions and individuals accepting the goals of high quality ontology in both theory and practice.
The mission of the Center for Computational Research (CCR) is to: (1) enable research and scholarship at UB by providing faculty with access to high-performance computing and visualization resources, (2) provide education, outreach, and training in Western New York, and (3) foster economic development and job creation in Western New York by providing local industry with access to advanced computing resources, including hardware, software and consulting services.In addition to providing access to state-of-the-art computational and visualization resources, CCR's staff, who are comprised of computational scientists, software engineers, and database administrators, provide a wide range of services to facilitate faculty led research including custom software and GUI interface development, advanced database design, scientific programming support, scientific visualization support, and bioinformatics support. In addition, CCR offers courses in high-performance computing and molecular modeling, training and workshops in specific aspects of high-end computing, a graduate certificate in computational science, and summer workshops for high-school students.
CCR also supports the newly established New York State Center of Excellence in Bioinformatics and Life Sciences, which is comprised of UB, the Roswell Park Comprehensive Cancer Center, and the Hauptman-Woodward Medical Research Institute.
The Intelligent Natural Language Processor (iNLP)
The iNLP processor is a DotNet architecture based general natural language processor (See Figure 1). The iNLP solution provides the highest accuracy available on the market or in any peer reviewed publication to date. For clinical problems the software has a sensitivity (i.e. Recall) of 99.7% and a positive predictive value (i.e. Precision) of 99.8%. This means that per 1,000 indexed concepts you will have on average only three false negatives and two false positives. This is more than an order of magnitude better performance than its closest competition.
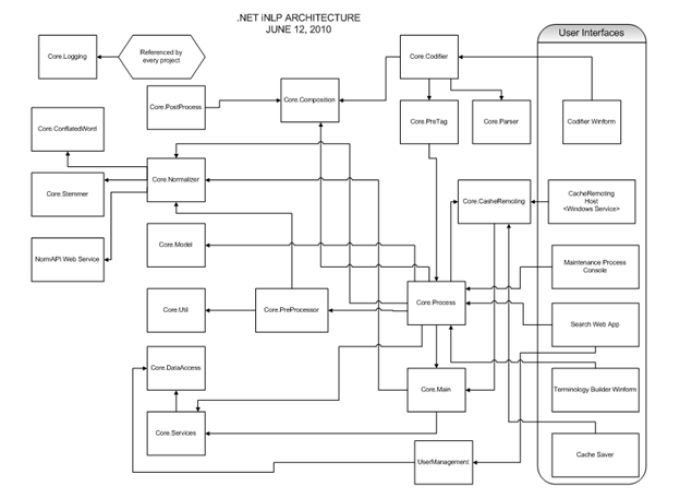
iNLP .Net Architecture Diagram.
The iNLP solution turns your free text data into structured data by assigning code values from national and internationally standard code sets (e.g. SNOMED CT, ICD9, ICD10, CPT, RxNorm, LOINC, Gene Ontology, etc.), thereby supporting interoperability.
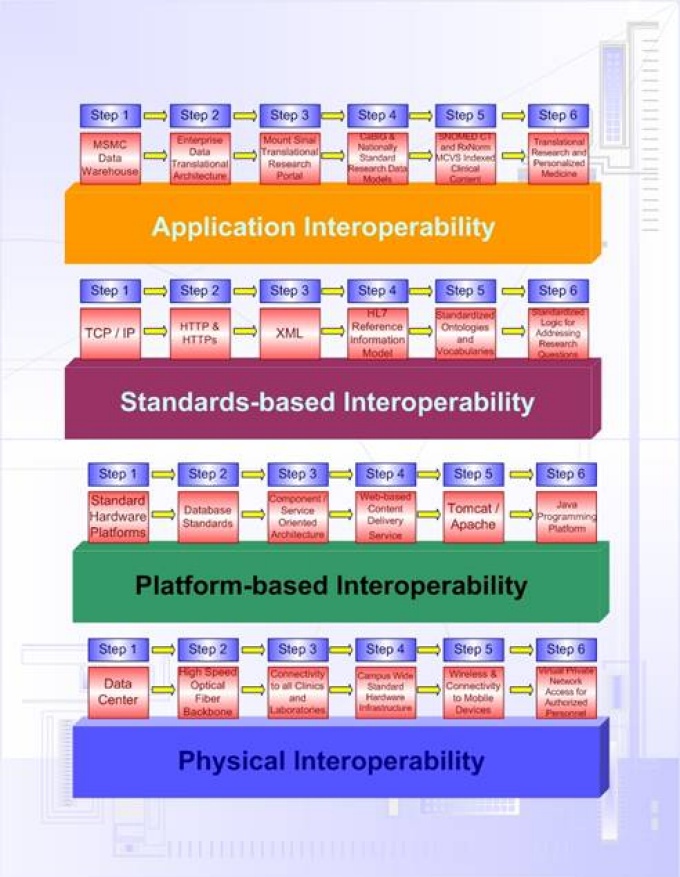
Interoperability guides the development of the iNLP resources to be conformant with National Health Informatics standards.
The solution is terminology independent so terminology servers can be built using your local code sets to support your personal business needs. Compositional expressions supporting post-coordination are generated automatically by the system.
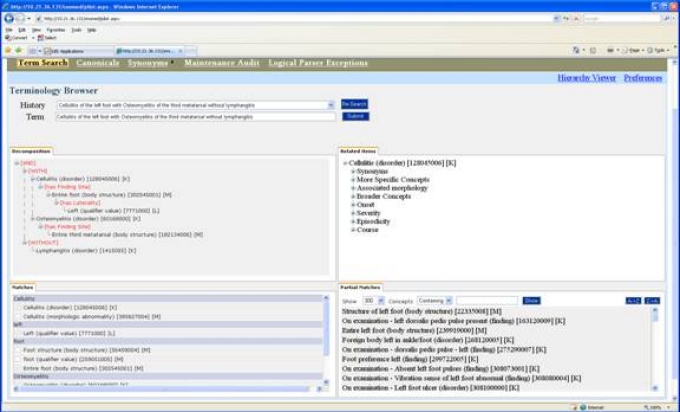
The iNLP terminology browser shows how the iNLP system generates logical compositional expressions.
The values and units available in the text are abstracted and associated with the concepts in the text (e.g. The patient’s total cholesterol was 245 mg/dl). The iNLP system has a temporal model that is designed to assign a start and end date to the concepts abstracted from the text. These concepts are tagged as either a positive, negative or uncertain assertion so that you can know the difference between facts that are true and facts that are false or are still being ruled in or out. This is critical for accurate and appropriate clinical decision support. This ontologically codified data is necessary for most all secondary uses of clinical data. This includes but is not limited to, firing clinical decision support rules, data driven recruitment to clinical trials, automated electronic quality monitoring, computer assisted or generated coding of both inpatient and outpatient encounters for Professional fee billing, post-market surveillance of drugs, management of the clinical practice, and to support rapid (10 minute) retrospective clinical trials.
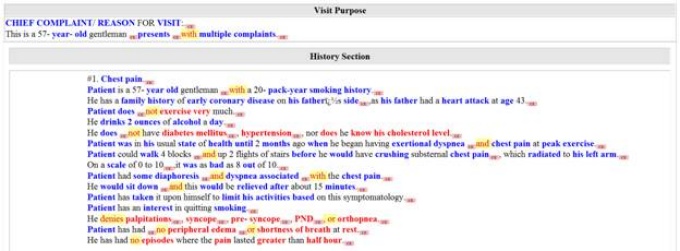
The iNLP fully encoded clinical record. Blue concepts are positive assertions, red are negative assertions and green concepts are uncertain assertions.
Our eQuality workbench produces validated quality rules to satisfy the meaningful use criteria, allowing healthcare organizations to capture additional funding from the Centers for Medicare and Medicaid Services. Here subject matter experts simply craft the rules in plain English and the system automatically codes the criteria to ensure sensitive and specific results. This process supports healthcare organizations’ continuous quality improvement programs.
The eQuality / eStudy workbench. This software allows simultaneous access to both structured and unstructured data. This frees subject matter experts to concentrate on their content while they create computable quality rules and study criteria.
Our eStudy workbench facilitates a researchers ability to produce computable inclusion / exclusion criteria for their studies and then provides a method for them to write the critical questions for each study and to even run the study, regularly receiving the study results in less than a minute. This makes research more approachable for all clinicians and researchers and provides them with the capability of modeling prospective trials as well as establishing preliminary data for future clinical trials.
Technically, the iNLP software is rock-stable and runs rapidly as a web service under IIS in the .Net framework. The system uses a .Net cache to provide real time indexing of clinical data and rapid retrieval times for aggregate queries such as electronic clinical trials.
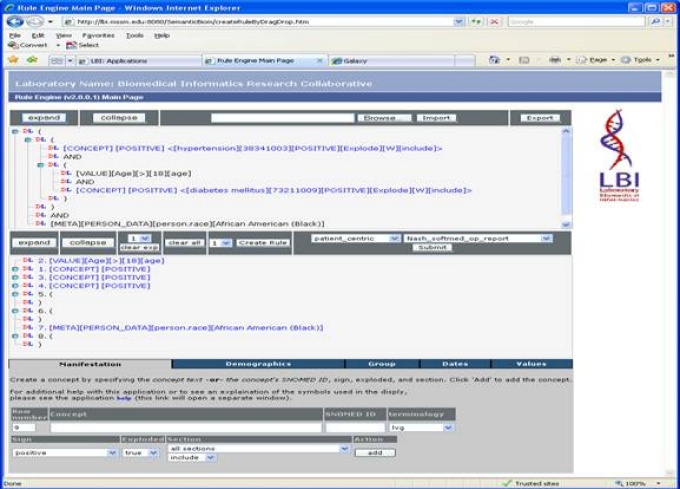
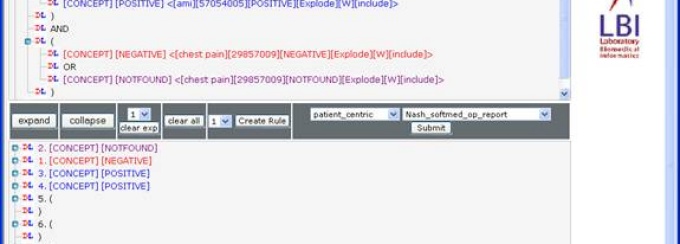
The iNLP eStudy workbench, looking at Hypertensive Diabetics over the age of 18 that are of African American heritage.
Aim: To provide as an Ontology service a natural language processing web service to Ontologically codify research data
Dr. Elkin and Dr. Ceusters have a long history of producing accurate NLP solutions. Language and Computing and Nuance’s NLP solutions are derived from Dr. Ceusters work and Cerner’s NLP and Semantic Search and Discern nCode is derived from Dr. Elkin’s work. Dr. Elkin has written permission from Mount Sinai to use the NLP software created at Mount Sinai in this research effort.
The software has a sensitivity (recall) of 99.7% and a positive predictive value (precision) of 99.8% and a specificity of 97.9% for clinical problems. The software has been validated for the creation of clinical quality rules, surveillance of post-operative complications and for biosurveillance.
We will create an Ontology server for each Ontology of interest in this project. These will be exposed via a web service that will be available to our internal research team, to our fellow cooperative agreement awardees and then once validated to the broader NIH funded research community.
The NLP web service’s WSDL will allow the researcher to choose the Ontology to use to codify the submitted research data. Data will be returned to the researcher for storage in a standards based common data infrastructure and further analysis. Multi-centered trails will be accomplished via distributed queries against this common data infrastructure. Queries such as the example of studies inclusion criteria (see Figure 6 below) can be created, saved and reused or distributed. Thus one could save separately the inclusion and exclusion criteria for a study and then for the critical question of interest. Then combine them and save the result separately.
Semantic Distance
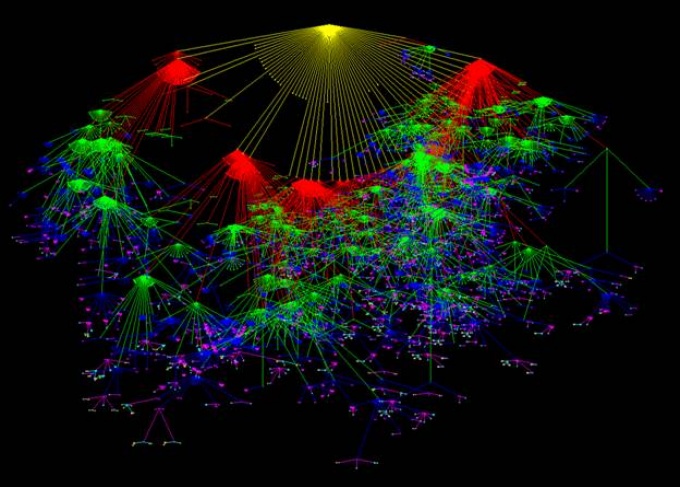
The formal conceptual distance between concepts in the SNOMED CT Cancer hierarchy. Each color is a layer, each node a disorder and each line the formal conceptual distance between parent and child.
In a recent study, semantic clustering was capable of clustering 17,180 of the 27,335 human genes while hierarchical clustering clustered only 5,553 of the human genes (p<0.001). The sensitivity (recall) of the semantic clustering was 99.5% while the sensitivity of the hierarchical clustering was only 66.7% (p<0.001). This experiment shows clearly the superiority of semantic clustering over hierarchical clustering. The superiority is in large part due to the ability to measure when entities are not equivalent how far apart they are in conceptual ontological distance. These shades of gray (rather than simply being able to tell black and white) represent a significant portion of the data with which researchers are faced. Semantic clustering will provide researchers with an important method with which they can approach big data clustering, discovery and analysis.
As this approach is dependent on the quality of the ontologies being employed having high quality ontologies available to the research community is essential for our efforts to handle, organize, classify and analyze big data. High quality ontologies allow us to create the electron microscope for looking at big data.
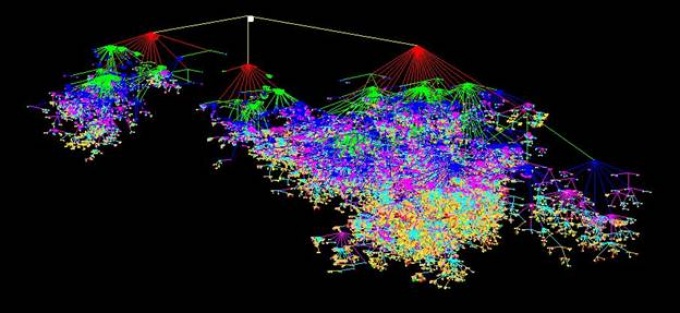
The Gene Ontology (GO) showing the formal conceptual distance between concepts. Each color is a level in the hierarchy. Each node is an entity in GO and the lines are the formal distance between concepts.
Example 1
Most human cancers show perturbation of growth regulation mediated by the tumour-suppressor proteins retinoblastoma (RB) and p53 (ref. 1), indicating that loss of both pathways is necessary for tumour development. Loss of RB function leads to abnormal proliferation related to the deregulation of the E2F transcription factors, but also results in the activation of p53, which suppresses cell growth. Here we show that E2F-1 directly activates expression of the human tumour-suppressor protein p14ARF (the mouse homologue is called p19ARF), which binds to the MDM2-p53 complex and prevents p53 degradation2,5. These results complete a pathway linking abnormal proliferative signals, such as loss of RB, with the activation of a p53 response, through E2F-1 and p14ARF. They suggest that E2F-1, a protein inherently activated by cell-cycle progression, is part of a fail-safe mechanism to protect against aberrant cell growth.
The blue entities are important elements of signaling and regulatory pathways and the red are functions linking entities within a pathway.
Semantic Triples Generated:
- growth regulation mediated by tumour-suppressor proteins retinoblastoma
- growth regulation mediated by p53
- Loss of RB function leads to abnormal proliferation
- abnormal proliferation deregulation of E2F transcription factors
- Loss of RB function leads to activation of p53
- Loss of RB function suppresses cell growth
- p53 suppresses cell growth
- E2F-1 activates expression human tumour-suppressor protein p14ARF
- human tumour-suppressor protein p14ARF binds to MDM2-p53 complex
- human tumour-suppressor protein p14ARF prevents p53 degradation
- loss of RB activation of p53 response
- p53 response through E2F-1
- p53 response through p14ARF
- E2F-1 activated by cell-cycle progression
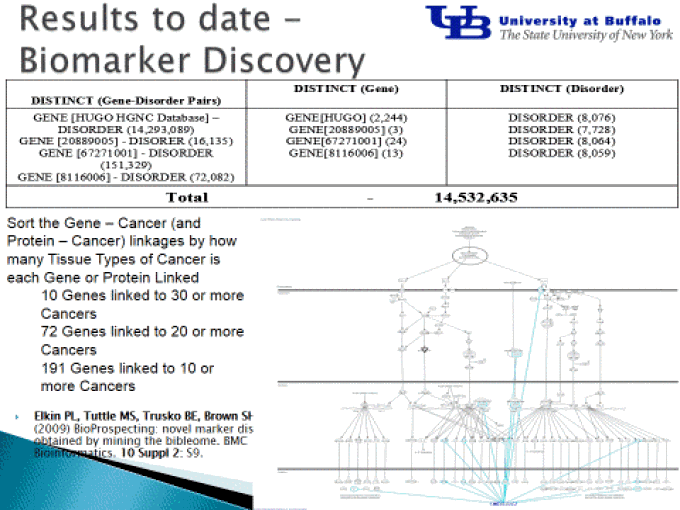
Text mining was shown to yield new relevant biological knowledge

Text mining the literature was used to suggest new systems biological signaling mechanisms
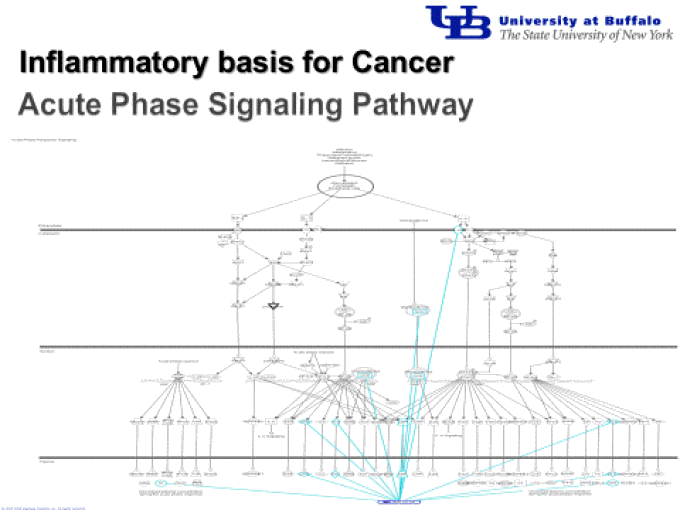
Mapping text mining results to the known regulome.
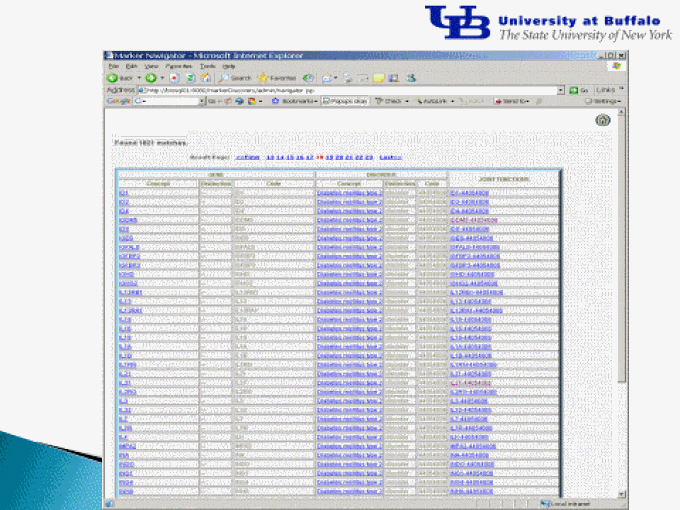
Results of the literature text mining shown in a web interface
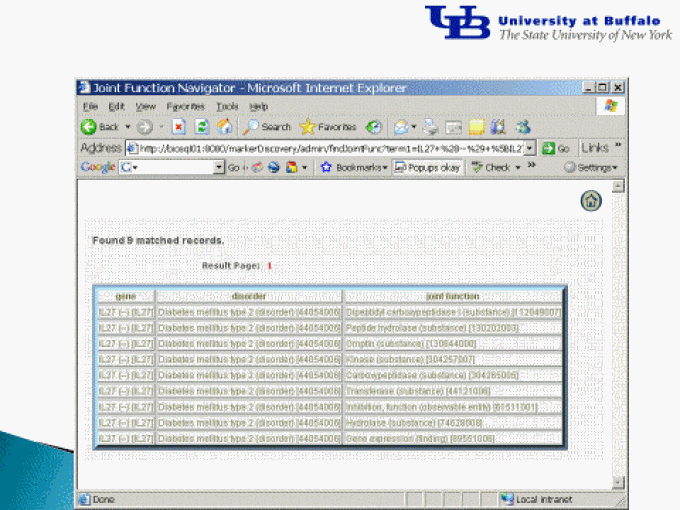
Examples of signaling relationships linking genes and disorders